Bacterial Staining:
- Negative Staining
- Smear Preparation and Simple Staining
- Gram Stain
- Acid-Fast Staining (Ziehl-Neelsen and Kinyoun) Procedure
- Endospore Staining (Schaeffer-Fulton or Wirtz-Conklin)
- Capsule Staining
- Flagella Staining: West and Difco’s SpotTest Methods
Culture Techniques:
- Sterilization and Microbiological Culture Media Preparation
- Culture Transfer Instruments, Techniques, and Isolation and Maintenance of Pure Cultures
- Spread-Plate Technique
- The Steak-Plate Technique: Selective and Differential Media
- Pour-Plate Technique
- Cultivation of Anaerobic Bacteria
- Determination of Bacterial Numbers
Biochemical Activities of Bacteria:
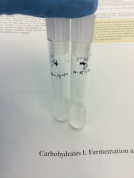
- Carbohydrates I: Fermentation and β-Galactosidase Activity
- Carbohydrates II: Triple Sugar Iron Agar Test
- Carbohydrates III: Starch Hydrolysis
- Lipids: Lipid Hydrolysis
- Proteins, Amino Acids, and Enzymes I: The IMViC Tests
- Proteins, Amino Acids, and Enzymes II: Casein Hydrolysis
- Proteins, Amino Acids, and Enzymes III: Gelatin Hydrolysis
- Proteins, Amino Acids, and Enzymes IV: Catalase Activity
- Proteins, Amino Acids, and Enzymes V: Coagulase and DNase Activity
- Proteins, Amino Acids, and Enzymes VI: Oxidase Test and Bioluminescence
- Proteins, Amino Acids, and Enzymes VII: Oxidase Test and Bioluminescence
- Proteins, Amino Acids, and Enzymes VIII: Urease Activity
- Proteins, Amino Acids, and Enzymes IX: Lysine and Ornithine Decarboxylase Tests
- Proteins, Amino Acids, and Enzymes X: Phenylalanine Deamination
- Proteins, Amino Acids, and Enzymes XI: Nitrate Reduction
Rapid Multitest Systems:
- The API® 20E System
- The Enterotube® II System
Unknown project strategy: Candidates
- Gram Positive: Bacillus cereus, B. megaterium, B. subtilis, Enterococcus faecalis, Listeria monocytogenes, Micrococcus luteus, Staphylococcus aureus.
- Gram Negative: Citrobacter freundii, Enterobacter aerogenes, Escherichia coli, Klebsiella pneumoniae, Proteus vulgaris, Pseudomonas aeruginosa, Salmonella arizonae, Shigella sonnei.
Identification flow charts Here NOTES S. marcescens produces red colonies B. subtilis produces white to cram colonies E. coli produces white colonies Blood agar is selective and differential media.
NOTES S. marcescens produces red colonies B. subtilis produces white to cram colonies E. coli produces white colonies Blood agar is selective and differential media.
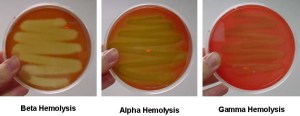
- It distinguishes between hemolytic and non-hemolytic bacteria.
- Streptococci and staphylococci are hemolytic.
- Gram positive results 11-15-14: No hemolytic bacteria


- Gram negative results 11-15-14: Hemolytic bacteria (beta?)
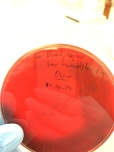
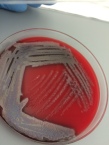
- Hemolytic bacteria produces clear zones around their colonies because of blood cell destruction.
Mannitol salt agar.
- Use to isolate staphylococci
- 7.5% sodium chloride inhibits the growth of most bacteria.
- Staphylococcus aureus will ferment the mannitol and form yellow zones in the reddish agar because phenol red becomes yellow in the presence of fermentation acids.
- Gram positive fermentation of acids:
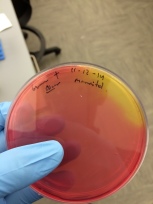 11-12-14
11-12-14 11-15-14
11-15-14
Eosin methylene blue agar (EMB agar).
- Largely used for the detection of E. coli. Lactose fermenters like E. coli will take up the dyes and form green to blue-black colonies with a metallic sheen
- It contains the dyes eosin Y and methylene blue, which partially suppress the growth of gram-positive bacteria.
- Lactose nonfermenters such as Salmonella, Proteus, Pseudomonas form colorless to amber colonies.
- Gram positive results 11-15-14

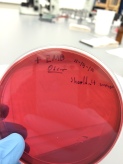 Little or non bacteria culture
Little or non bacteria culture - Gram negative results 11-15-14

 Growth but not E. coli, no green-sheen.
Growth but not E. coli, no green-sheen.
Deep agar for cultivating anaerobic bacteria.
- C. sporogenes is an obligate anaerobe.
- E. coli is a facultative anaerobe. It will grow either aerobically or in the absence of oxygen, but better in its presence.
- P. aeruginosa is a strict aerobe.
Durham tube for fermentation activity and ONPG for beta-Galactosidase activity.
- E. coli produces acid and gas by catabolizing D-glucose (dextrose) and other carboxydrates.
- E. coli is beta-Galactosidase positive since it can also use lactose as its sole source of carbon.
- Alcaligenes faecalis is an obligately aerobic rod, coccal rod, or coccus that uses acetate, propionate, butyrate, and some other organic acids as a sole carbon and energy source; because carbohydrates are not used, it does not produce acid or gas.
- Salmonella cholerae-suis is a facultatively anaerobic gram-negative rod having both a respiratory and fermentative type of metabolism, it produces acid and sometimes gas. It is beta-Galactosidase positive.
- Gram positive results:


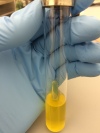 Acidity but not gas production.
Acidity but not gas production. - Gram negative results:


 No acidity, no gas.
No acidity, no gas. - ONPG negative in Gram positive and Gram negative, results:
Triple sugar iron (TRI) agar test to differentiate among the family of Enterobacteriaceae.
- eg of enterobacteriaceae: E. coli, Proteus vulgaris, and Shigella flexneri.
- All three from the eg are gram negative rods, facultative anaerobic.
- E. coli produces an acid butt, an acid or alkaline slant, and no H2S, but does produce gas.
- P. vulgaris produces an acid butt, an acid or alkaline slant, H2S, and gas.
- S. flexneri produces an acid butt, an alkaline slant, no H2S, and no gas.
- A. faecalis produces an alkaline butt, alkaline slant, no H2S, and no gas.
- P. aeruginosa produces an acid butt, alkaline slant, no H2S, and no gas.
- Gram positive results 11-15-14:

 facultative anaerobe, yellow butt and slant. No gas H2S formation.
facultative anaerobe, yellow butt and slant. No gas H2S formation. - Gram negative results 11-15-14:

 strict aerobe, red butt and slant. No gas H2S formation. The bacteria is not from the Enterobacteriaceae.
strict aerobe, red butt and slant. No gas H2S formation. The bacteria is not from the Enterobacteriaceae.
- Yellow batt and red slant due only to the fermentation of glucose. The slant remains red because of the limited glucose in the medium and, therefore, limited acid formation, which does not persist since the oxidative decarboxylation forms alkaline amines that increase the pH.
- A yellow butt and acid slant due to the fermentation of glucose and lactose and/or sucrose leads to excessive acid formation in the entire medium.
- Gas formation noted by splitting or lifting of the agar.
- Gas formation (H2S) seen by blackening of the agar.
- Red butt and slant indicate that none of the sugars were fermented and neither gas nor H2S were produced.
Starch Hydrolysis for the presence of hydrolases.
- If alpha-amylase is not produced, the bacterium will not hydrolyze starch.
- Bacillus subtilis is amylase positive.
- E. coli is amylase negative.
- Proteus vulgaris is variable, it maybe positive or negative.
- Results:

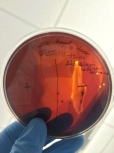 11-15-14 Gram – is hydrolase positive, gram + is hydrolase negative.
11-15-14 Gram – is hydrolase positive, gram + is hydrolase negative.
IMViC series of tests distinguish between different enteric bacteria.
- Indole test for the presence of indole due to tryptophanase activity
- Methyl red test for acidic intensity and pH
- Voges-Proskauer for the presence of precursor acetoin on glucose fermentation
- Citrate test for the use of citrate as energy source
- Enterobacter aerogenes is a facultatively anaerobic gram negative rod that has peitrichous flagella. It is a motile lactose fermenter. It is indole negative, MR negative, VP positive, and Simmons citrate positive.
- E. coli is a facultatively anaerobic gram-negative rod that is motile with peritrichous flagella or nonmotile. It is a lactose fermenter. It is indole positive, MR positive, VP negative, and Simmons citrate negative.
- Klebsiella oxytoca is a facultatively anaerobic gram-negative rod. It is nonmotile and a lactose fermenter. It is indole positive, often MR negative, VP positive, and Simmons citrate positive.
- Proteus vulgaris is a gram-negative anaerobic rod. It has peritrichous flagella, is motile, and does not ferment lactose. It is indole positive, MR positive, VP negative, and sometimes Simmons citrate positive.
Indole Production
- Bacteria that contain the enzyme tryptophanase can hydrolyze tryptophan to its metabolic products, namely, indole, pyruvic acid, and ammonia. The bacteria use the pyruvic acid, and ammonia to satisfy nutritional needs; indole is not used and accumulates in the medium.
- The presence of indole can be detected by the addition of Kovacs’ reagent. It reacts with the indole, producing a bright red compound on the surface of the medium.
- Results:

 11-16-14 Gram – is positive and Gram + is negative for the presence of tryptophanase.
11-16-14 Gram – is positive and Gram + is negative for the presence of tryptophanase.
Methyl Red Test
- All enteric bacteria catabolize glucose for their energy needs; however the end products vary depending on the enzyme pathways present in the bacteria. The pH indicator methyl red tects a pH change to the acid range as a result of acidic end products such as lactic, acetic, and formic acids. It distinguishes between E. coli (mixed acid fermenter) and E. aerogenes (a butanediol fermenter).
- The pH of the medium does not fall as low as during mixed acid fermentation. At a pH of 4, the methyl red indicator turns red, it is a positive methyl red test. At a pH of 6, the indicator turns yellow, a negative test.
Voges-Proskauer Test
- It identifies bacteria that ferment glucose, leading to 2,3-butanediol accumulation in the medium. The addition of 40% KOH and 5% solution of alphanaphthol in absolute ethanol(Barritt’s reagent) will detect the presence of acetoin, a precursor in the synthesis of 2,3-butanediol. In the presence of the reagents and acetoin, a cherry-red color develops. Develop of this color after 15 minutes represent a positive test.
- Results: 11-16-15
 Gram + is positive and Gram – is negative for the presence of acetoin in glucose fermentation
Gram + is positive and Gram – is negative for the presence of acetoin in glucose fermentation
Citrate Utilization Test
- It determines the ability of bacteria to use citrate as a sole carbon source for their energy needs. This ability depends on the presence of a citrate permease that facilitates transport of citrate into the bacterium. Once inside the bacterium, citrate is converted to pyruvic acid and CO2. Simmons citrate agar slants contain sodium citrate as the only carbon source, NH4+ as a nitrogen source, and the pH indicator bromotrymol blue. When bacteria oxidize citrate, they remove it from the medium and liberate CO2. CO2 combine with sodium and water to form sodium carbonate, an alkaline product. This raises the pH , turns the pH indicator to a blue color and represents a positive citrate test.
- Results 11-15-14:

 Gram – is positive and Gram + is negative for the presence of citrate permease for citrate utilization.
Gram – is positive and Gram + is negative for the presence of citrate permease for citrate utilization.
Catalase activity test for destruction of hydrogen peroxide on aerobic bacteria
- aureus is a gram-positive coccus that is catalase positive when grown in an aerobic environment. It is mainly associated with the human skin and mucous membranes of warm-blood vertebrates, but is often isolated from food products, dust, and water.
- E. faecalis is a catasae-negative, gram-positive coccus.
- Micrococcus luteus is another gram-positive coccus that also is catalase positive.
- Reduction of O2 produces superoxide radicals and hydrogen peroxide, or the hydroxyl radical. Those are extremely toxic because they are powerful oxidizing agents and destroy cellular constituents very rapidly. For protection against this, obligate aerobes and facultative anaerobes usually contain the enzyme superoxide dismutase, which catalyzes the destruction of superoxide, and either catalase or peroxidase, which catalyzes the destruction of hydrogen peroxide.
- Most strict anaerobes lack both enzymes and therefore cannot tolerate O2. If catalase was produced by the bacteria, it will be liberated O2.
- Results 11-16-14 (mistake using plates instead of slant)

 gram – is positive and gram + is negative for the presence of catalase to destroy the hydrogen peroxide.
gram – is positive and gram + is negative for the presence of catalase to destroy the hydrogen peroxide.
Coagulase Test for the presence of Staphylococci
- Coagulase-producing staphylococci form a fibrin clot around themselves and avoid attack by the host’s defenses.
- A positive test will cause the plasma to clot by using coagulase to initiate the clotting cascade.
- Results:
 Gram – is positive and Gram + is negative for the presence of coagulase, charasteristic of staphylococci.
Gram – is positive and Gram + is negative for the presence of coagulase, charasteristic of staphylococci.
Oxidase Test
- Alcaligenes faecalis is a gram-negative, aerobic rod that possesses a strictly respiratory type of metabolism with oxygen as the terminal electron acceptor. It is thus oxidase positive.
- This test distinguishes between groups of bacteria based on cytochrome c oxidase activity.
- E. coli is a facultatively anaerobic gram-negative rod that has both respiratory and fermentative types of metabolism and is oxidase negative.
- P. aeruginosa is a gram-negative, aerobic rod having a strictly respiratory type of metabolism with oxygen as the terminal electron acceptor and thus oxidase positive.
- The ability of bacteria to produce cytochrome oxidase can be determined by the addition of the oxidase text reagent or test strip. Oxidase positive will turn blue.
- Results: (mistake using slant instead of plates)
 Gram – is positive and gram + is negative for the presence of cytochrome oxidase (for oxidation of reduced cytochrome c and water formation) in aerobic respiration.
Gram – is positive and gram + is negative for the presence of cytochrome oxidase (for oxidation of reduced cytochrome c and water formation) in aerobic respiration.